Method for the computation of bootstrapped confidence intervals.
Usage
confidence_intervals(
data,
pred_model,
h,
train = 0.8,
M = 1000,
alpha = 0.05,
output = NULL,
...
)Arguments
- data
Data.frame (or vector) to apply cross-validation where the rows are the observations and the columns are the variables.
- pred_model
Model to predict the data where the first argument in the data and the second the length of prediction. Additional parameters can be passed using ... . Returns a vector of the predictions.
- h
Number of observations to predict into the future.
- train
Minimum amount of data (0,1) to train model. After this point, a sliding window approach is taken.
- M
Numeric. Number of bootstrap iterations.
- alpha
Significance level for the intervals
- output
Numeric or name of the column for the output when using a data.frame for
data. If not given, the first column is used.- ...
Additional parameters to pass to
pred_model.
Examples
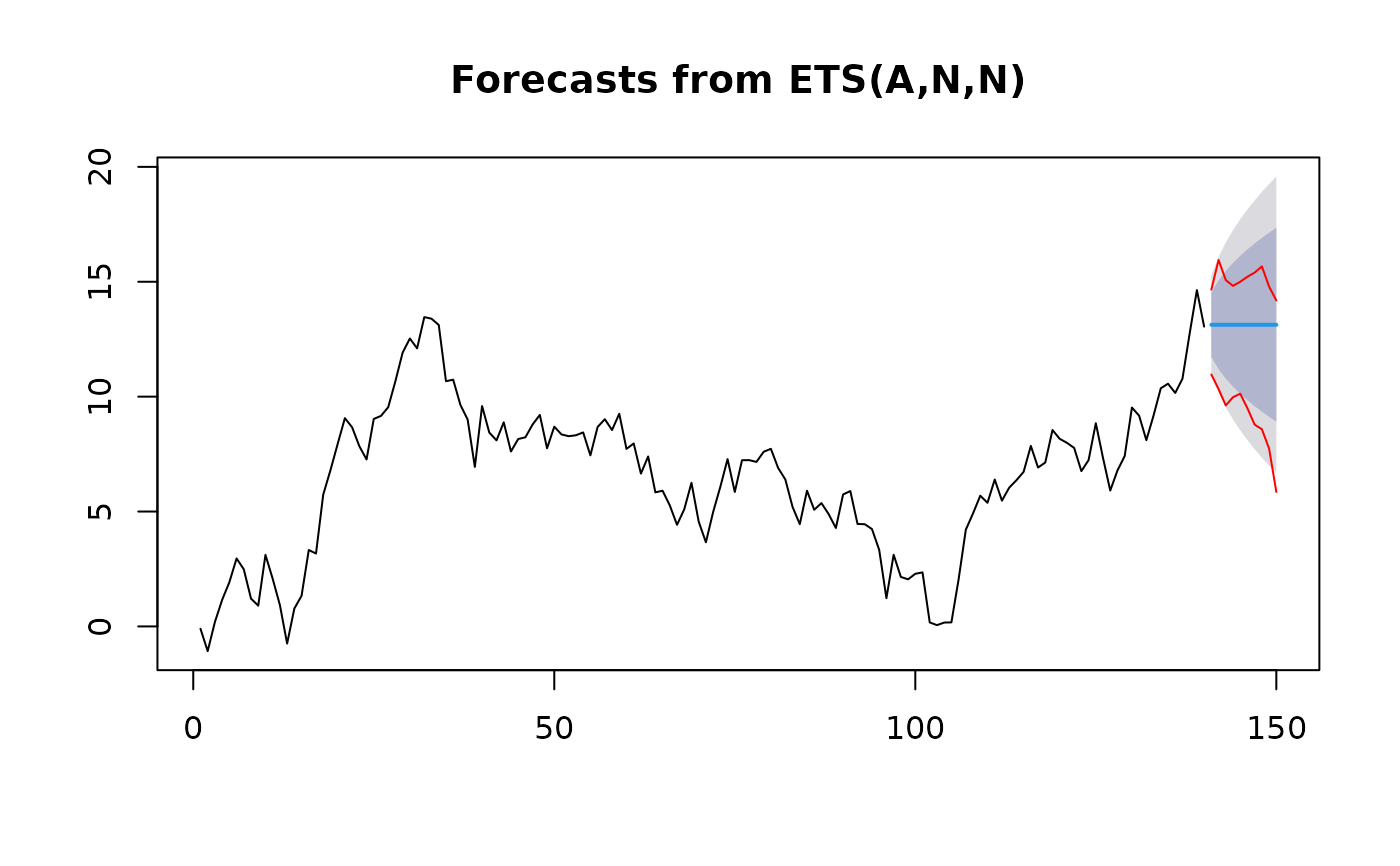
data <- cumsum(rnorm(150))
pred_model <- function(x, h) {
predict(forecast::ets(x), h = h)$mean
}
h <- 10
ets_model <- predict(forecast::ets(data[1:140]), h)
#> Registered S3 method overwritten by 'quantmod':
#> method from
#> as.zoo.data.frame zoo
ints <- confidence_intervals(data = data[1:140], pred_model = pred_model, h = h)
plot(ets_model)
lines(x = 141:150, y = ints$lower, col = "red")
lines(x = 141:150, y = ints$upper, col = "red")